ElGamacy Protein Design Lab
Our protein design team develops and deploys cutting-edge design methods to generate novel proteins for biomedical applications. Our group's research lies at the interface of protein design, protein biophysics, and translational medicine. At present, we are actively researching the following topics:
Development of high-performance tools for computational protein designs
The task of calculating the energy associated with a given structure lies at the core of protein design. Improving the accuracy and speed of energy calculations can enable us to design more complex proteins more efficiently. Our group works on developing tools at the bleeding edge of protein design methodology. These tools are under active development:
- The Damietta tensorized protein design engine (published)
- The Protein Design Toolkit (published)
- HECTOR: A pipeline for ultra-fast docking and de novo binder design (published)
Accelerating molecular dynamics
Computing atomic motions is central to understanding and predicting the properties of
proteins, as well as other materials.
Our group is currently
researching more efficient methods to calculate interatomic
forces and accurately simulate their motions.
Design of therapeutic signaling modulators
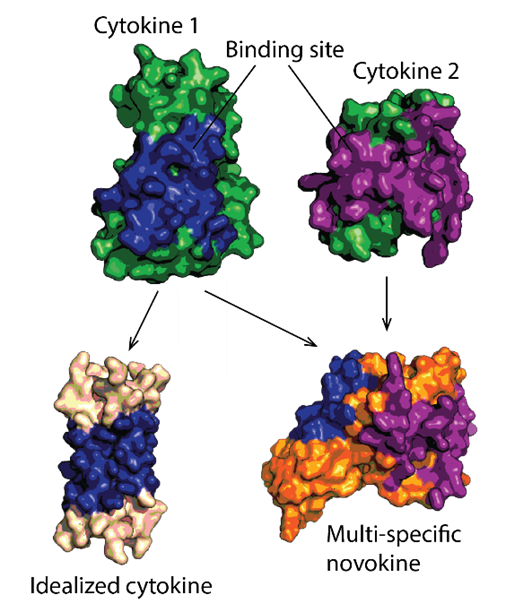
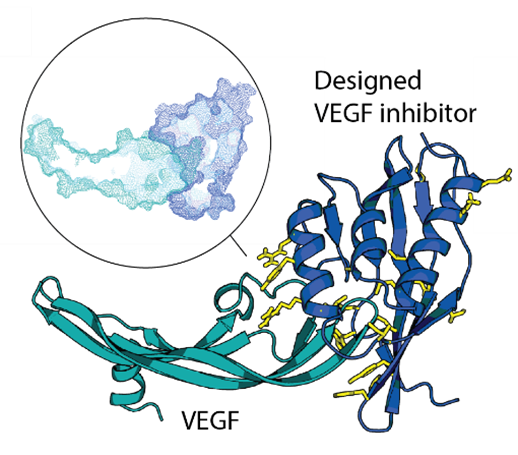
Protein therapeutics must meet a range of requirements to their activity, stability, solubility, aggregation propensity, and production costs. Computational design can offer control over these parameters and greatly facilitate protein drug development. We use a set of in-house developed computational methods to make idealized therapeutic miniproteins. We design and preclinically-develop idealised proteins to modulate cytokine and growth factor signaling by targeting receptors or their ligands. We also use these designs to study the structure-function relationship of receptor modulation.
Design of synthetic proteins
By extending our physics-based modeling and design software infrastructure, we are currently working on designing and validating synthetic proteins. These proteins are built from non-canonical amino acids and designed to possess superior therapeutic properties to proteins made from the 20 canonical amino acids.